Photosynthesis is not the only dioxygen-evolving biological process. For instance, chlorite dismutase (Cld; EC 1.13.11.49) catalyses the production of O2 from chlorite (1):
| ClO2− → Cl− + O2 | (1) |
The reaction (1) is not really disproportionation, and NC-IUBMB made a valid point that the term “chlorite dismutase” is “misleading”. Even so, the NC-IUBMB-approved, sorry, “accepted” name “chlorite O2-lyase” for an oxidoreductase is equally absurd; I am going to ignore it.
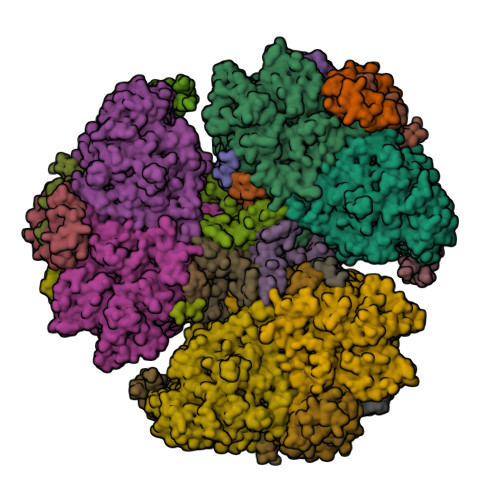
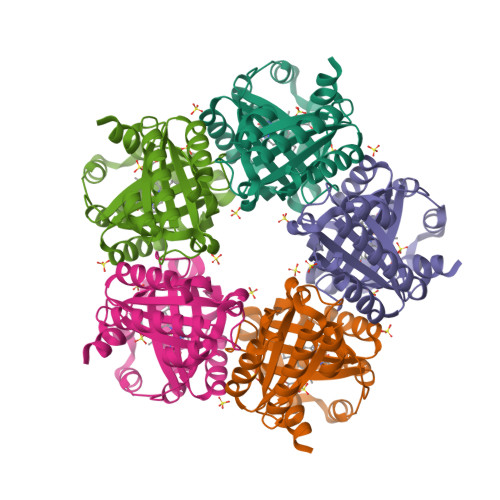
Chlorite dismutase from Azospira oryzae exists as a homohexamer [1] while Cld from Dechloromonas aromatica [2] and enzyme from Candidatus Nitrospira defluvii [3] are homopentamers.
 |  |
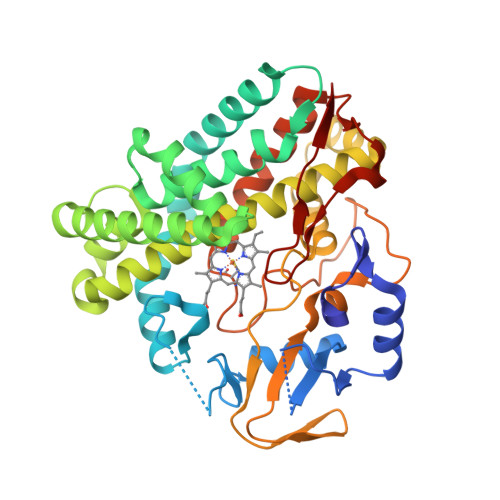
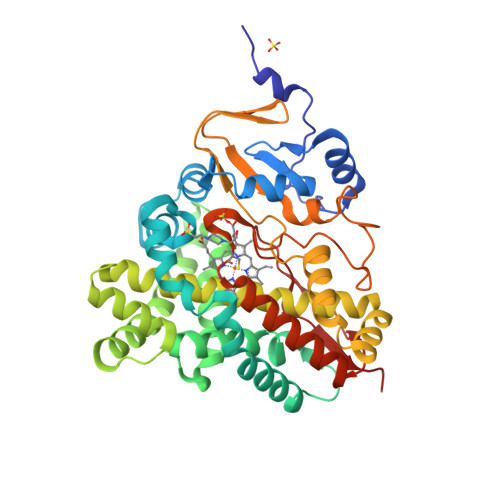
The active site contains a single haem group [Fe(ppIX)] coordinated by a proximal histidine residue. Goblirsch et al. [2] propose the mechanism where the reaction of chlorite within the distal pocket of Cld generates hypochlorite (ClO−) and a compound I intermediate [Fe(ppIX)O] (1a). Then ClO− rebounds with compound I forming the chloride and dioxygen (1b):
| ClO2− + [Fe(ppIX)] → ClO− + [Fe(ppIX)O] | (1a) |
| ClO− + [Fe(ppIX)O] → Cl− + O2 + [Fe(ppIX)] | (1b) |
- de Geus, D.C., Thomassen, E.A.J., Hagedoorn, P.-L., Pannu, N.S., van Duijn, E. and Abrahams, J.P. (2009) Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 387, 192—206.
- Goblirsch, B.R., Streit, B.R., DuBois, J.L. and Wilmot, C.W. (2010) Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 15, 879—888.
- Kostan, J., Sjöblom, B., Maixner, F., Mlynek, G., Furtmüller, P.G., Obinger, C., Wagner, M., Daims, H. and Djinović-Carugo, K. (2010) Structural and functional characterisation of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: Identification of a catalytically important amino acid residue. J. Struct. Biol. 172, 331—342.