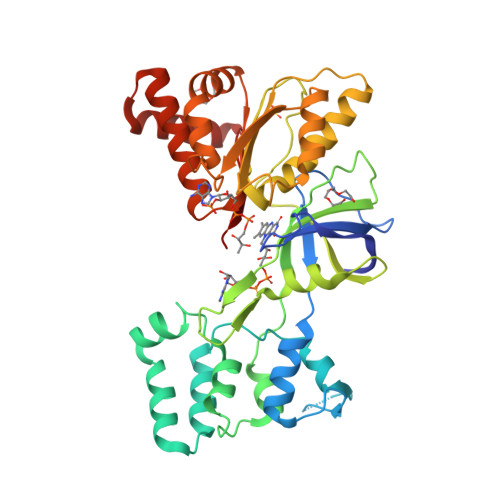
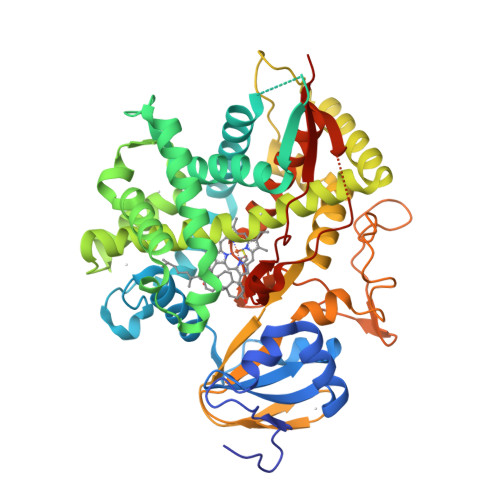
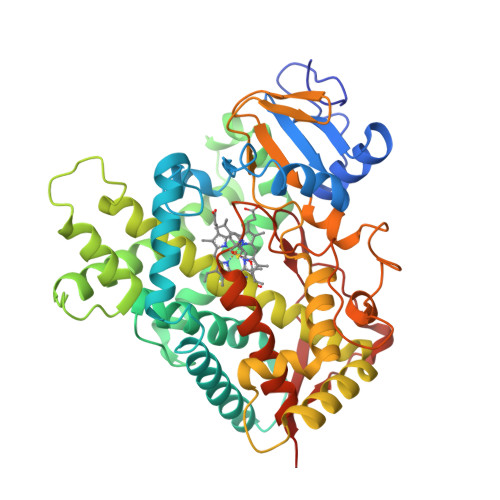
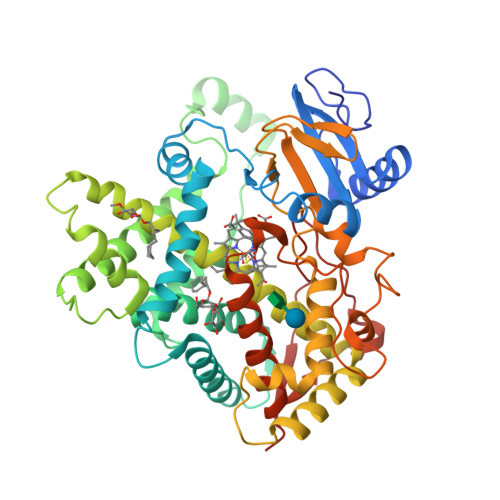
The epsilonproteobacterium Wolinella succinogenes is able to grow by sulphite respiration with formate as electron donor [1], thanks to the octahaem cytochrome c MccA that catalyses the six-electron reduction of sulphite to sulphide:
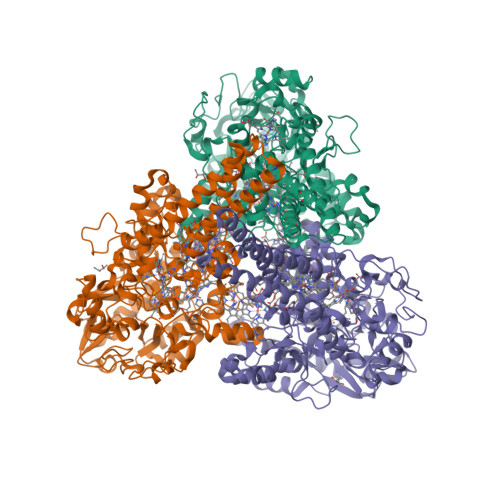
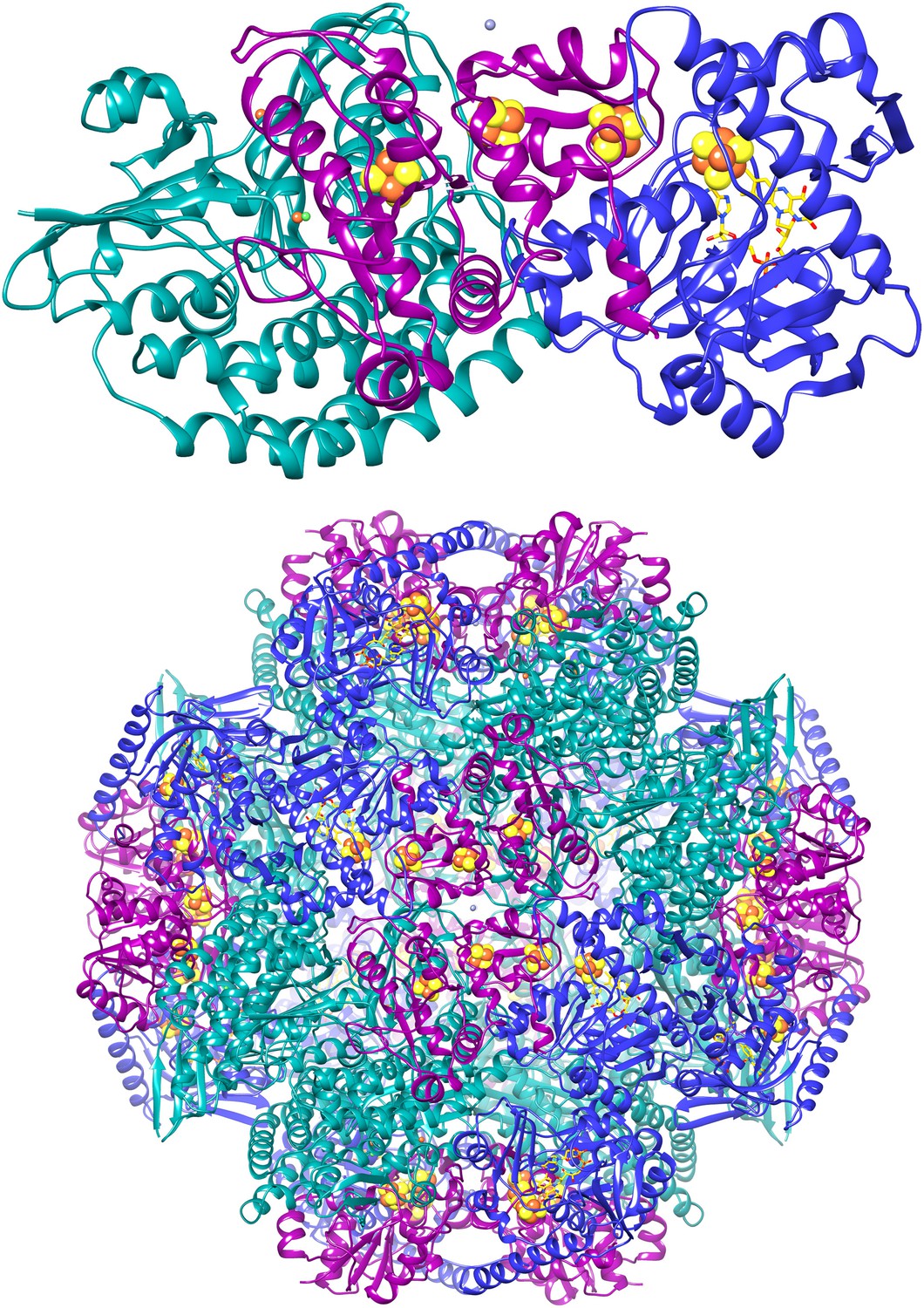
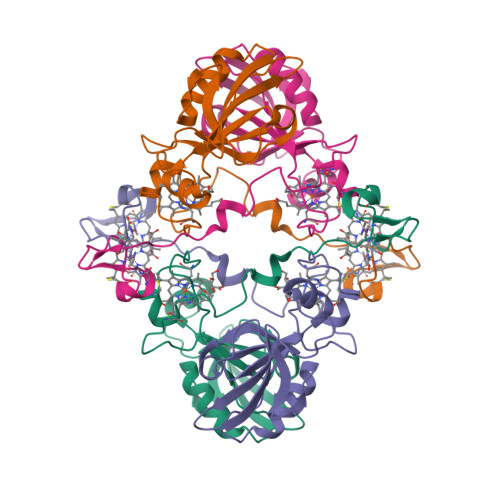
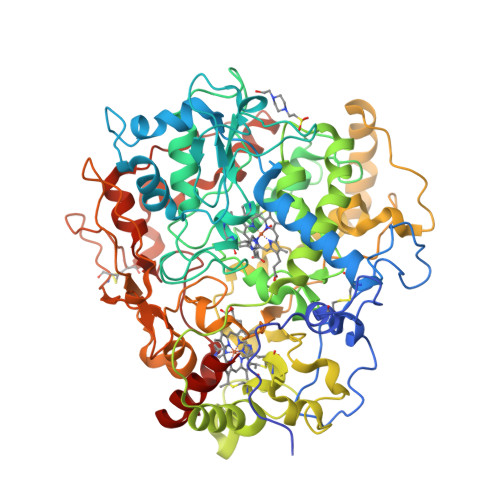
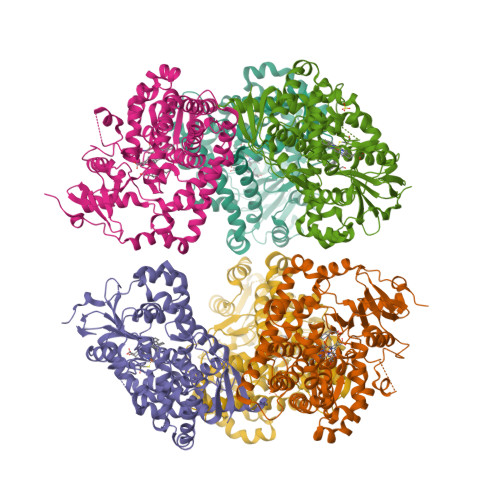
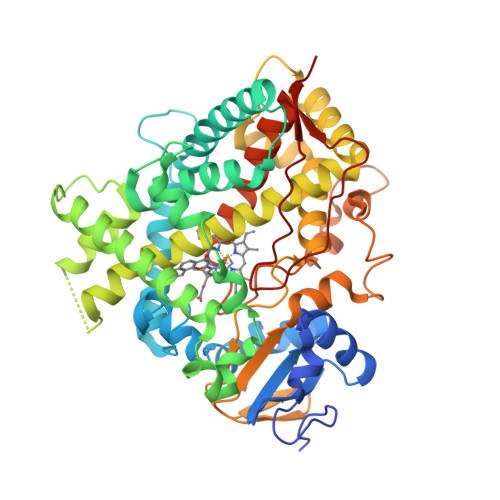
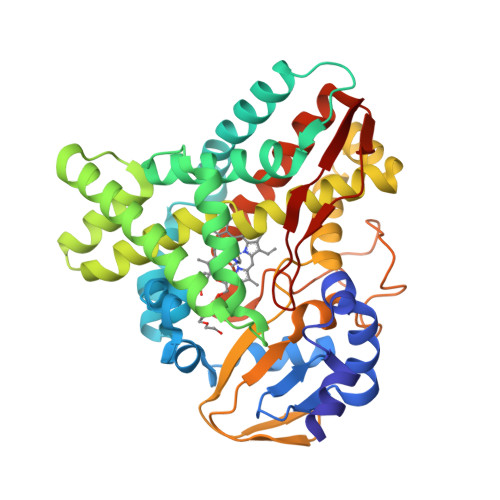
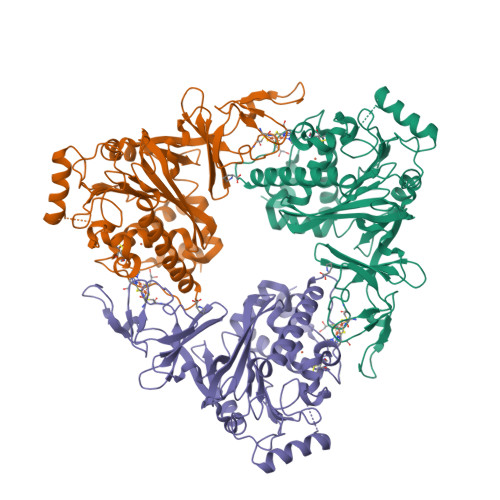
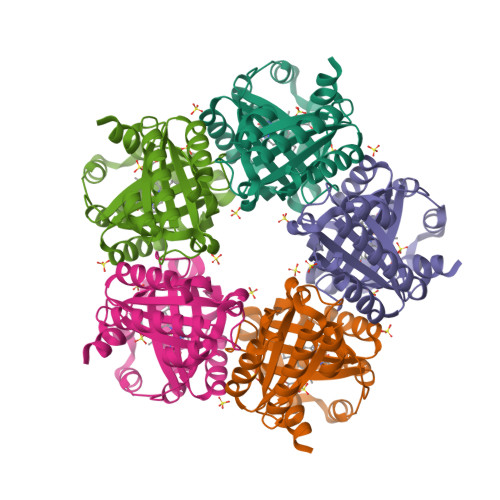
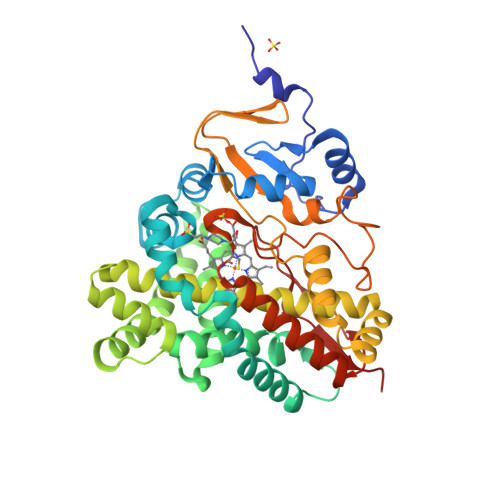
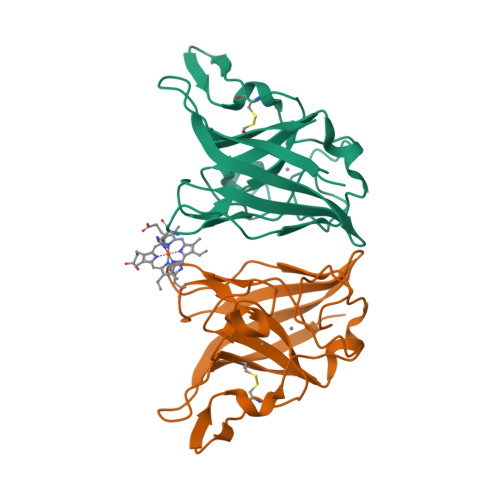
The crystal structure of MccA has been determined at 2.2 Å resolution [2, 3]. The enzyme exists as a homotrimer showing a novel fold and haem arrangement. The heterobimetallic active centre contains a Cu(I) ion and a haem c with a Fe—Cu distance of 4.4 Å [4].
|
- Kern, M., Klotz, M.G. and Simon, J. (2011) The Wolinella succinogenes mcc gene cluster encodes an unconventional respiratory sulphite reduction system. Molecular Microbiology 82, 1515—1530.
- PDB:4RKM
- PDB:4RKN
- Hermann, B., Kern, M., La Pietra, L., Simon, J., Einsle, O. (2015) The octahaem MccA is a haem c-copper sulfite reductase. Nature 520, 706—709.