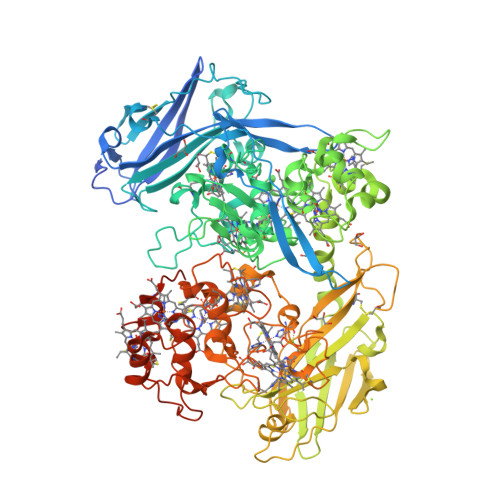
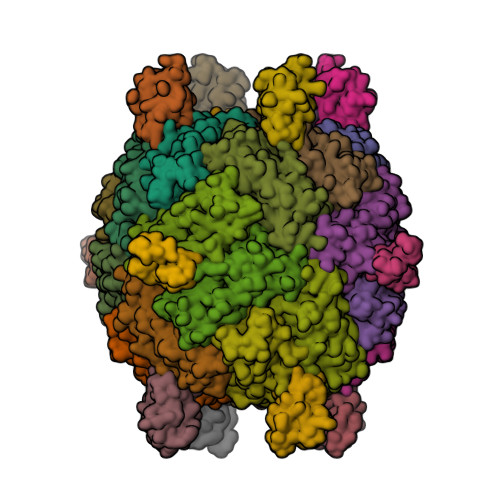
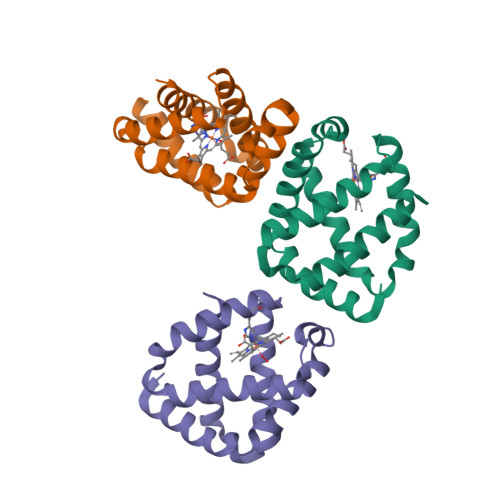
This year I don’t have a Christmas tree. (There aren’t many in Fuerteventura.) But I’ve got some models from this week’s new PDB structures [1—5] which look Christmassy enough to decorate my blog. Merry Christmas and a happy New Year everybody!

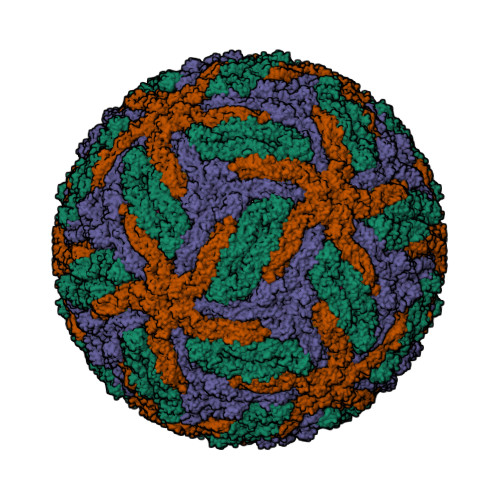
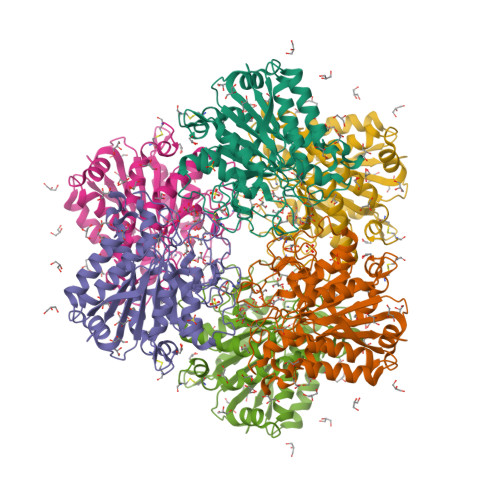
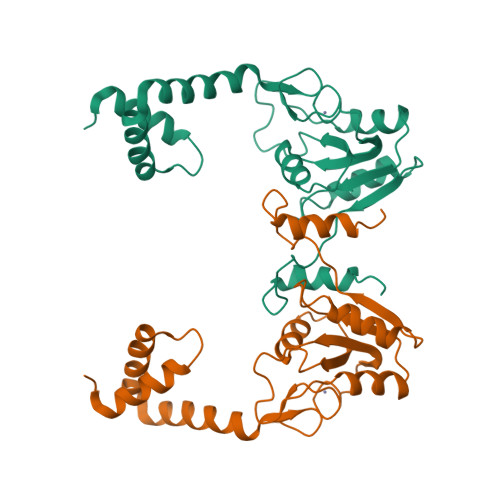
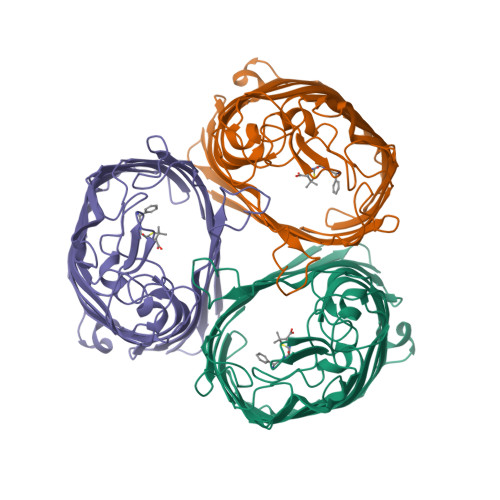
- Hui, F., Scheib, U., Hu, Y., Sommer, R.J., Aroian, R.V. and Ghosh, P. (2012) Structure and glycolipid binding properties of the nematicidal protein Cry5B. Biochemistry 51, 9911—9921.
- Lence, E., Tizón, L., Otero, J.M., Peón, A., Prazeres, V.F.V., Llamas-Saiz, A.L., Fox, G.C., van Raaij, M.J., Lamb, H., Hawkins, A.R. and González-Bello, C. (2013) Mechanistic basis of the inhibition of type II dehydroquinase by (2S)- and (2R)-2-benzyl-3-dehydroquinic acids. ACS Chem. Biol. 8, 568—577.
- Strugatsky, D., McNulty, R., Munson, K., Chen, C.-K., Soltis, S.M., Sachs, G. and Luecke, H. (2013) Structure of the proton-gated urea channel from the gastric pathogen Helicobacter pylori. Nature 493, 255—258.
- Tang, Q., Gao, P., Liu, Y.-P., Gao, A., An, X.-M., Liu, S., Yan, X.-X. and Liang, D.-C. (2012) RecOR complex including RecR N-N dimer and RecO monomer displays a high affinity for ssDNA. Nucleic Acids Res. 40, 11115—11125.
- Ziervogel, B.K. and Roux, B. (2013) The binding of antibiotics in OmpF porin. Structure 21, 76—87.